NanoPhotometer®: Small Volume Spectrophotometric Applications in Human Genetics
NanoPhotometer® Application Leaflets
Small Volume Spectrophotometric Applications in
HUMAN GENETICS
Congenital defects caused by submicroscopic copy number variants (CNVs)
Introduction:
Congenital defects like limb malformations and intellectual disabilities are relatively common in the human population, affecting 3-5% of newborns. In the great majority of cases the origin is unknown. To investigate the underlying genetic causes, screening approaches such as microarraybased comparative genomic hybridization (array CGH) have been shown to be successful tools. We applied array CGH as the initial screening tool to detect copy number variations (CNVs) i.e. gains (duplications) and losses (deletions) in patient’s genomic DNA. In addition, besides research projects, array CGH has in recent years become an important tool in human genetic diagnosis of patients presenting with intellectual disability, development delay and/or congenital malformations. High quality DNA is absolutely mandatory to achieve good results. Therefore we used the multifunctional NanoPhotometer® from Implen with its unique optical path, to precisely quantify on the one hand the genomic DNA prior to labelling, and on the other hand to quantify and control the labelling efficiency prior to hybridization. With its high precision in small sample volume analysis the NanoPhotometer® is best suited for state of the art applications such as array CGH.
METHODS
Array CGH:
Patient’s and reference DNA (1 µg each) were differentially labelled with fluorescent dyes (Cy3 and Cy5, respectively) by random priming reaction and subsequently hybridized to an oligonucleotide array. Prior to labelling the DNA concentration was quantified by the Implen NanoPhotometer®, measuring the absorbance at 260 nm (1.5 µl sample volume). The purity and integrity was assessed by the 260/280 ratio (protein impurity) and agarose gel electrophoresis of the sample. Following the random priming
labelling reaction and purification, each sample was again analysed by the NanoPhotometer® to ensure equal amounts of dye incorporation in patient and reference samples (Figure 1). The small volume option was highly appreciated since we avoid wasting valuable labelled samples. Preprogrammed applications on the NanoPhotometer® allow measurement of Frequency of Incorporation (FOI). Thus, the instrument enables an easy, fast and reliable analysis of the labelling reaction.
Quantitative Real-Time PCR (qPCR)
To validate any potential pathogenic changes detected by array CGH analysis we employed quantitative Real-Time PCR (qPCR). Specific primers were designed to amplify fragments located within the region of interest. Factor VIII served as internal control to determine the patient’s sex. Genomic DNA was measured in small volume using the NanoPhotometer® and subsequently diluted to a concentration of 2 ng/µl. qPCR was performed as described in detail by Klopocki et al. 2008. Relative quantification of copy number was performed using the standard curve method as described by the manufacturer. AACt (cycle threshold) values were calculated by comparing the Ct value of samples with the copy number control albumin (ALB), followed by normalization to the calibrator (normal female DNA).
RESULTS:
Measurement of Frequency of Incorporation (FOI)
By determining the FOI we were able to adjust the amounts of labelled DNA between patient and reference to an equal amount. This allows for better results and less normalization efforts in the subsequent array CGH experiment (Figure 1).
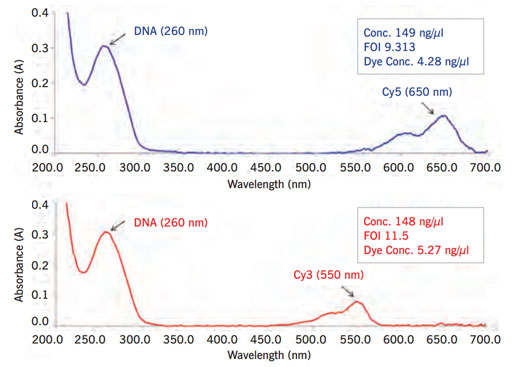
Figure 1: Measurement of labelling efficiency with NanoPhotometer® in patient (Cy3, upper panel) and reference (Cy5, lower panel) samples at 550 nm (Cy3) and 650 nm (Cy5).
Microduplications cause congenital limb malformations
Our array CGH analyses detected submicroscopic duplications at the SHH locus on chromosome 7q36 (Figure 2A). associated with polydactyly phenotype in patients (title photos), (Klopocki et al. 2008).
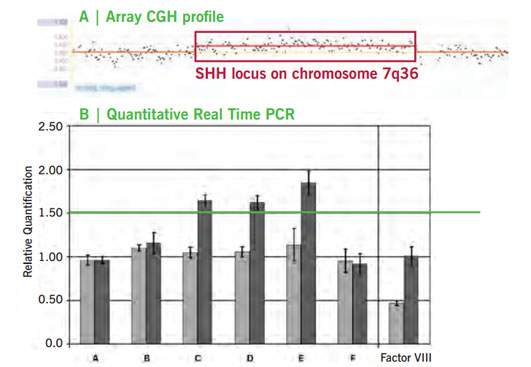
Figure 2: Microduplication on chromosome 7q36 associated with polydactyly. (A) Array-CGH profile of SHH locus. Duplicated region indicated by the red box (log2ratio > 0.4). (B) Confirmation of microduplication byqPCR. Amplicons C-E show increased copy number i.e. relative quantification > 1.5 in affected (dark grey) compared to controls (light grey). Phenotypic spectrum with extra finger on the preaxial (thumb) side or finger-like thumb is presented on the title photo.
The array CGH results were successfully confirmed by qPCR (Figure 2B). SHH is a morphogen important in embryonic development. The expression of SHH during development is controlled by various noncoding regulatory elements upstream of SHH. The detected duplication affects an element known as ZRS (ZAP regulatory sequence) which significantly induces SHH expression in the limb bud. Duplications as well as point mutations of the ZRS lead to ectopic expression of SHH in the limb bud and thus cause the observed polydactyly phenotype.
Detection of novel microdeletion syndromes in patients with developmental delay, intellectual disability and epilepsy
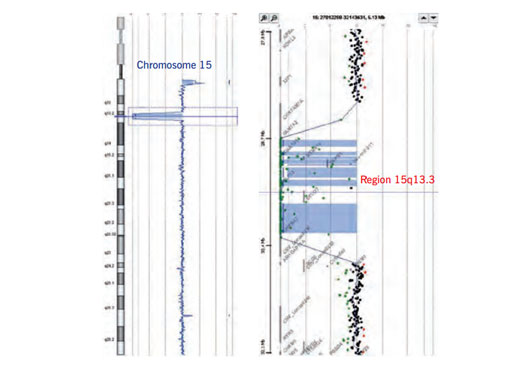
One of the major benefits of array CGH compared to conventional cytogenetics (chromosome analysis) is the ability to detect submicroscopic aberrations i.e. aberrations smaller than 5 Mb. This enables the identification of novel microdeletion and microduplication syndromes. We detected a homozygous deletion of chromosome 15q13.3 in two severely affected sibs presenting with intellectual disability, development delay, no speech development, seizures, muscular hypotonia, and congenital malformations of the eyes (Spielmann et al. 2011).One of these is microdeletion on 15q13.3 encompassing the CHRNA7 gene (Figure 3). Patients with heterozygous deletions present with developmental delay (DD), intellectual disability (ID), facial dysmorphism and epilepsy. In case of a homozygous deletion indicated by log2ratio of -4 (Figure 3) the phenotype is more severe.
References: Klopocki E, Ott C-E, Benatar N, Ullmann R, Mundlos S, Lehmann K. (2008) A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. Journal of Medical Genetics 45(6):370-5.
Spielmann M, Reichelt G, Hertzberg C, Trimborn M, Mundlos S, Horn D, Klopocki E. (2011) Homozygous deletion of chromosome 15q13.3 including CHRNA7 causes severe mental retardation, seizures, muscular hypotonia, and the loss of KLF13 and TRPM1 potentially cause macrocytosis and congenital retinal dysfunction in siblings. European Journal of Medical Genetics. 2011 Apr 29. [Epub ahead of print]
Summary:
Congenital malformations are relatively common in the human population affecting 3-5% of newborns. However, the underlying cause is often unknown. We were able to demonstrate an association between duplications at the SHH locus on the long arm of chromosome 7 and a hand malformation characterized by additional fi ngers (polydactyly) or fi nger-like thumbs (triphalangeal thumb). The second example shows the benefi t of high resolution array CGH in human genetic diagnosis. We detected the underlying genetic cause in two severely affected sibs presenting with intellectual disability, developmental delay, no speech development, seizures, muscular hypotonia, and congenital malformations of the eyes. Our fi ndings show that rare CNVs can cause a wide spectrum of congenital disease, ranging from intellectual disability to limb malformations. The small size of these CNVs (< 5 Mb) precludes detection by conventional methods such as karyotyping or chromosomal CGH. Thus, array CGH is nowadays an invaluable tool in both genetic research and human genetic diagnosis. For this project the Implen NanoPhotometer® , with its integrated submicroliter options, was successfully utilized for DNA measurements and determination of labeling effi ciency in the small volume application. Since amount of sample is often limited we highly appreciate the possibility of reliable small volume measurements. The wide application spectrum, ease of use, and fast, accurate performance of the recalibration-free NanoPhotometer® makes this instrument an important tool for daily laboratory work in human genetic resarch and diagnosis.
We would like to thank Dr. Eva Klopocki, Institute for Medical Genetics and Human Genetics, Charité – University Hospital Berlin, Germany, for the friendly offer of measuring data and the title photo, and for supporting us during compilation of the Human Genetics application leafl et!





