NanoPhotometer®: Small Volume Spectrophotometric Applications in Pathology
NanoPhotometer® Application Leaflets
Small Volume Spectrophotometric Applications in
PATHOLOGY
Individualized cancer diagnostics – mutation detection of EGF receptor and KRAS gene
Introduction:
For the past decades, treatment of cancer in humans has concentrated on three basic therapeutic options: surgery, chemotherapy and radiation. Although billions of dollars and manpower have been invested in searching for breakthroughs in new therapies, only now there is a growing awareness that the future in cancer treatment is based on individual therapies. Therefore, personalised diagnosis is needed. Two examples for this development are treatment of nonsmall cell lung cancer (NSCLC) with tyrosine kinase inhibitors (TKIs) and the treatment of metastatic colorectal carcinoma with monoclonal antibodies (mAB) like Cetuximab. The response rates to these drugs are very good, and the treatment is relatively well tolerated. But in the case of NSCLC, this holds true only for those patients that express an activating mutation in the epidermal growth factor receptor (EGFR) (Lynch et al., 2004). Likewise, treatment success of metastatic colorectal carcinoma with Cetuximab depends on a nonmutated KRAS gene (Di Fiore et al., 2007, Figure 1).
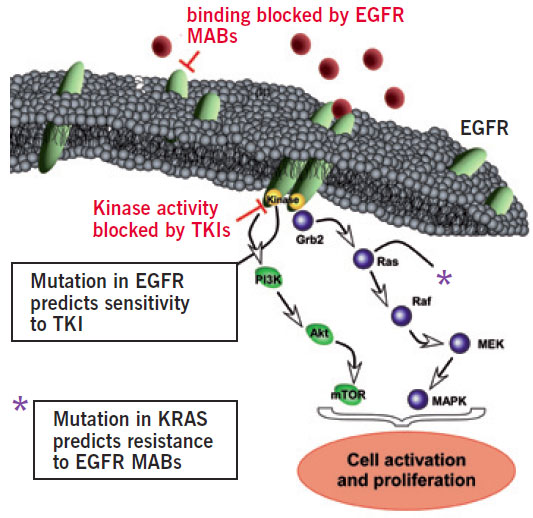
Figure 1: The EGFR/KRAS signalling cascade is switched on by activation of the external EGFR domain by circulating EGF, which causes release of downstream factors by the internal EGFR domain. The following cascade stimulates growth and division of the parent cell. This process is out-of-control in cancer patients and might be inactivated by TKIs or by mABs. Success of TKI therapy depends on an activating mutation of the EGFR. Mutation of the KRAS gene predicts resistance to mABs.
In order to benefit from TKI as well as from Cetuximab therapy, the patient’s tumour genome has to be analyzed for the existence of any mutation prior to therapeutic treatment. As the tumour material often consists of very small biopsy samples, accurate DNA quantification of the purified genomic DNA is a critical step. Here the Implen NanoPhotometer® with its easy, accurate performance and perfected analysis of very small sample volume was successfully applied for DNA quantification.
METHODS
DNA extraction and amplification
Tumour tissue samples originated from NSCLC and metastatic colorectal carcinoma patients. DNA from the tumour area was extracted as described by Zinsky et al. (2010). The DNA quantification at 260nm was quickly and efficiently afforded by small volume quantification of the genomic DNA with the Implen NanoPhotometer®. This instrument minimizes the effort for this routine laboratory work due to simple sample handling as well as high throughput rates, based on very short measuring times. After extraction and quantification of the genomic DNA, the EGFR exons 18, 19, 21 and the KRAS exon 2 were amplified by PCR (Lynch et al. 2004, Zinsky et al. 2010).
Mutation analysis
The analysis of EGFR exon 19 was done by direct capillary analysis of the PCR product. Sequencing analysis of EGFR exon 18 and KRAS exon 2 was performed as described by Lynch et al. 2004 and Zinsky et al. 2010. For fragment analysis of EGFR exon 21, the PCR product was digested with Msc I. SNaPshot analysis was performed with specific primers to codon 12 (Zinsky et al. 2010). The resulting fragments after sequencing, restriction digestion and SNaPshot analysis were finally analyzed by capillary electrophoresis.
RESULTS:
EGFR mutations
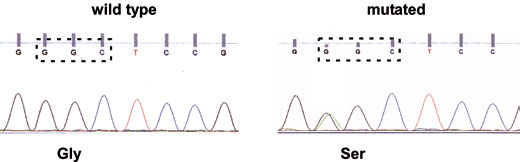
Using the described methodology, nearly 100% of all relevant mutational variants responsible for sensitivity to TKIs could be identified. Point mutation on exon 18 of the EGF receptor could be detected by sequencing analysis (Figure 2). In this case, a guanine to adenine mutation was found. This point mutation converts the amino acid (AA) glycine to serine, resulting in an activated EGFR and thus becoming sensitive to TKI therapy.
Figure 2: Sequence analysis of exon 18. Left side: wild type sequence coding the AA glycine. Right side: guanine to adenine point mutation changing the AA glycine to serine (arrow).
 
In frame deletions in exon 19 between 9 bp and 18 bp count for approximately 50% of all described EGFR mutations. Direct capillary analysis of the PCR product of exon 19 revealed an additional peak at 100 bp (arrow) in contrast to the wild type peak at 115 bp (Figure 3). This deletion of 15 bp in this part of the EGFR gene results in activation of EGFR, and therefore indicates treatment with TKIs.
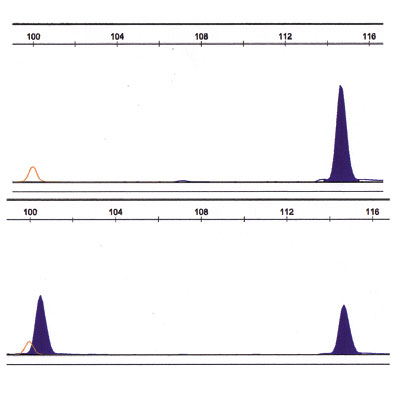
Figure 3: Fragment analysis of exon 19. Upper panel: wild type fragment (right blue peak at 115 bp). Lower panel: additional peak (left blue at 100 bp).

Figure 4: Fragment analysis of exon 21. Upper panel: wild type fragment at 43 bp (left filled blue peak). Lower panel: additional filled blue peak at 103 bp (arrow).
Another major mutation in the EGFR gene, accounting for about 40 % of all described mutations, is a point mutation in exon 21 at AA position 858. This mutation alters an intrinsic enzyme restriction site for Msc I, preventing the enzyme from cutting the respective PCR fragment. Fragment analysis of EGFR exon 21 after restriction digestion showed an additional peak at 103 bp (Figure 4, arrow). This longer fragment originates from undigested PCR product, indicating a point mutation in this part of the EGFR gene. This leads to activated EGFR and possible therapeutic success with TKIs.
KRAS gene mutations
In 45 out of 100 analyzed tissue samples, KRAS mutations in codon 12 or 13 were detected. 88.9% of these mutated KRAS genes were discovered by both DNA sequencing and SNaPshot, further 11.1% were only detectable by SNaPshot. Figure 5 shows mutation of codon 12 position 2 guanine to adenine transition. Mutation of the KRAS gene leads to an activated G-protein, even in absence of EGF, resulting in further malignant proliferation of the cells despite treatment (Di Fiore et al., 2007). Because of this, only patients with a nonmutated KRAS gene should be treated with Cetuximab.
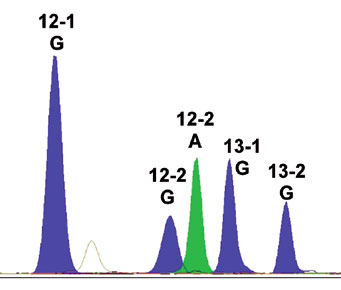
Figure 5: Electrophero- gram of the KRAS gene exon 12 (guanine to adenine transition) after SNaPshot analysis.
References:
Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., Louis D.N., Christiani D.C., Settleman J., Haber D.A., Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med, 2004; 350(21): 2129-2139. Di Fiore F., Blanchard F., Charbonnier F., Le Pessot F., Lamy A., Galais M.P., Bastit L., Killian A., Sesboüé R., Tuech J.J., Queuniet A.M., Paillot B., Sabourin J.C., Michot F., Michel P., Frebourg T., Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br. J Cancer, 2007; 96(8): 1166-1169. Zinsky R., Bölükbas S., Bartsch H., Schirren J., Fisseler-Eckhoff A., Analysis of KRAS Mutations of Exon 2 Codons 12 and 13 by SNaPshot Analysis in Comparison to Common DNA Sequencing. Gastroenterology Research and Practice, 2010, 2010:789363. Epub 2010 Dec 21.
SUMMARY:
Molecular cancer diagnostics facilitate individualized therapy for nonsmall cell lung cancer (NSCLC) and metastatic colorectal carcinoma patients. NSCLC patients profi t from therapy with tyrosine kinase inhibitors (TKIs) only if they express an activating mutation in the epidermal growth factor receptor (EGFR). Success of therapy of metastatic colorectal carcinoma with monoclonal antibodies depends on nonmutated KRAS gene. Therefore, molecular cancer characteristics strongly correlate with the response to a chosen chemotherapy. With sequencing, fragment and SNaPshot analysis, the presented data depict different ways to characterize the molecular structure of the cancer tissue.
In the majority of cases, individualized tumour diagnostics have to be performed with minimal amounts of genomic DNA from biopsy samples. Genomic DNA quantifi cation of the valuable tumour DNA is an essential prerequisite for subsequent mutation analysis. The Implen NanoPhotometer® accounts for accurate measuring results with very small sample volumes. In addition, due to the patented Sample Compression Technology™, the NanoPhotometer® is recalibration free and provides accurate measurements over the entire life-time of the instrument.
Further applications in Pathology / NanoPhotometer® reference customers:
Department of Pathology and Lab Medicine, Weill Cornell Medical College, New York, USA
“Measurements of nucleic acids, oligos and proteins in small volumes an OD600 of E. coli culture in standard cuvettes”
University Hospital of Bellvitge, Barcelona, Spain
“DNA quantifi cation in normal and paraffin tissues”
Medizinische Hochschule Hannover, Institute für Zell-und Molekularpathologie, Hannover, Germany
“Concentration determination of DNA and RNA for various array- applications, expression and CGH”
University of Münster, Germany
“Small volume concentration determination of RNA extracted from cell cultures and native tissues”